代表性图片
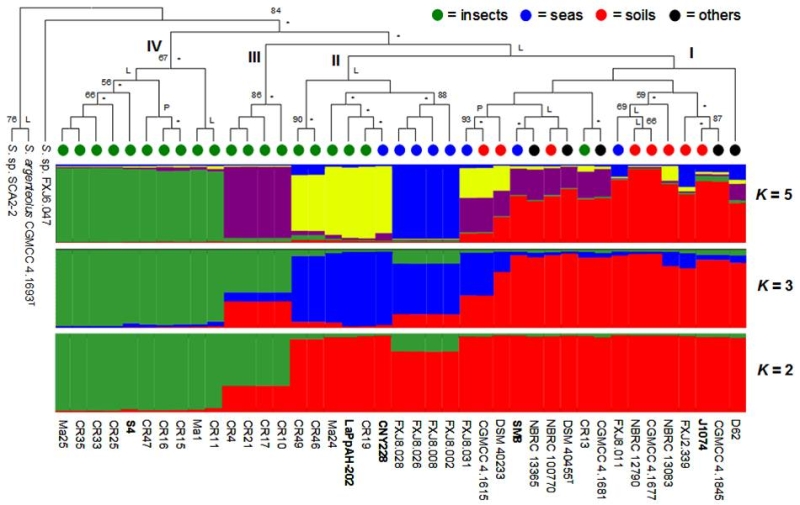
图1 微黄白链霉菌种群系统学与Structure分析
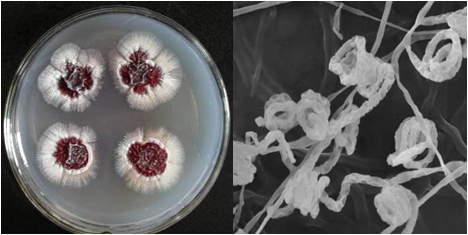
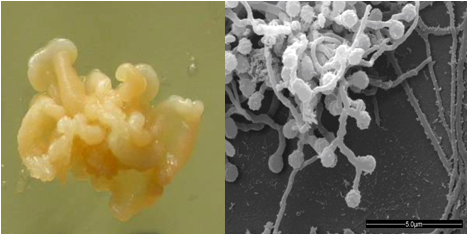
图2 链霉菌和小单孢菌的菌落及电镜形态
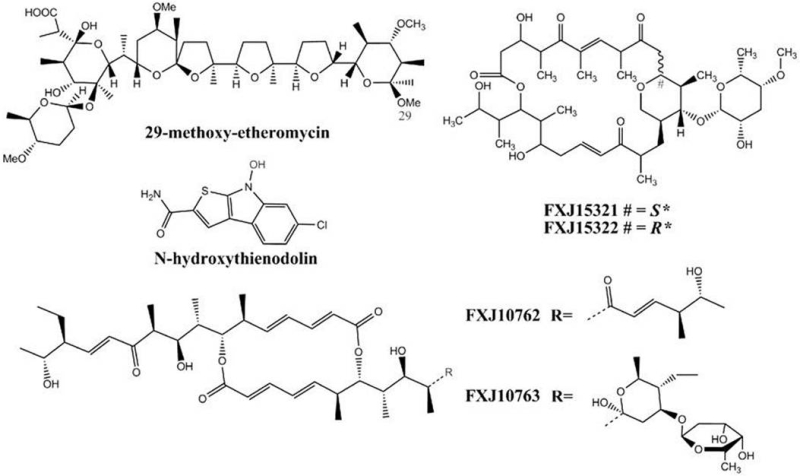
图3 红壤放线菌的部分活性代谢产物
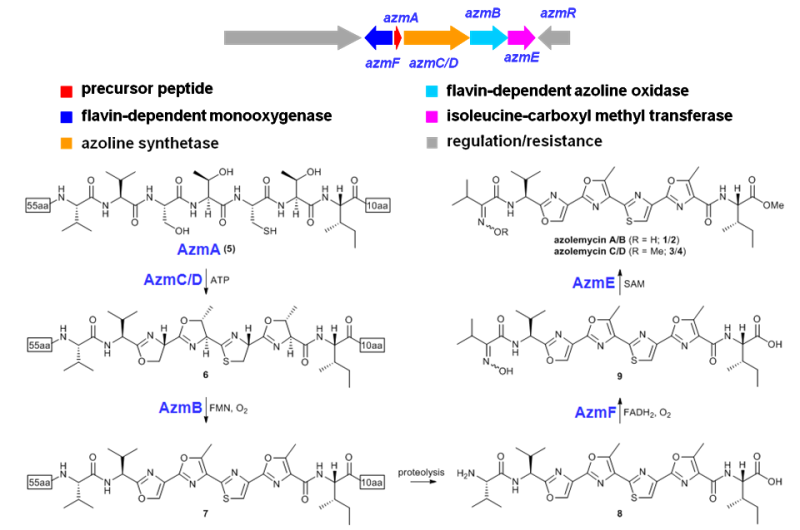
图4 Azolemycin生物合成途径解析
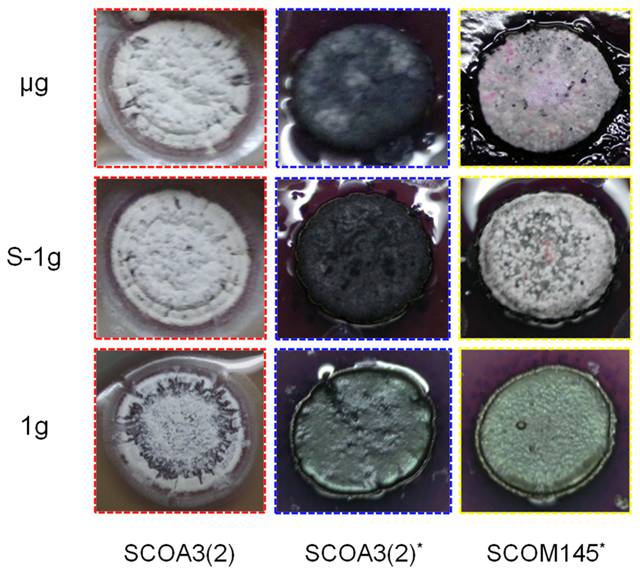
图5 模拟微重力及空间飞行促进链霉菌发育分化
代表性论文(#共同第一作者; *通讯作者):
1. Wang J, Li Y, Pinto-Tomás AA, Cheng K, Huang Y*. 2022. Habitat adaptation drives speciation of a Streptomyces species with distinct habitats and disparate geographic origins. mBio. https://doi.org/10.1128/mBio.02781-21
2. Li Y, Huang Y*. 2022. Distribution and evolutionary history of sialic acid catabolism in the phylumActinobacteria. Microbiol Spectr. https://doi.org/10.1128/Spectrum.02380-21
3. Yan B, Liu N, Liu M, Du X, Shang F, Huang Y*. 2021. Soil actinobacteria tend to have neutral interactions with other co-occurring microorganisms, especially under oligotrophic conditions. Environ Microbiol, 23(8): 4126–4140. https://doi.org/10.1111/1462-2920.15483
4. Zhang L, Chen Y, Xia Q, Kemner KM, Shen Y, O'Loughlin EJ, Pan Z, Wang Q, Huang Y, Dong H*, Boyanov MI*. 2021. Combined effects of Fe(III)-bearing clay minerals and organic ligands on U(VI) bioreduction and U(IV) speciation. Environ Sci Technol, 55(9):5929-5938. https://doi.org/10.1021/acs.est.0c08645
5. Yan B, Guo X, Liu M, Huang Y*. 2020. Ktedonosporobacter rubrisoli gen. nov., sp. nov., a novel representative of the class Ktedonobacteria, isolated from red soil, and proposal of Ktedonosporobacteraceae fam. nov. Int J Syst Evol Microbiol, 70:1015-1025. https://doi.org/10.1099/ijsem.0.003864
6. Zhang L, Zeng Q, Liu X, Chen P, Guo X, Ma LZ, Dong H*, Huang Y*. 2019. Iron reduction by diverse actinobacteria under oxic and pH-neutral conditions and the formation of secondary minerals. Chem Geol, 525: 390-399. https://doi.org/10.1016/j.chemgeo.2019.07.038
7. Li Y, Pinto-Tomás AA, Rong X, Cheng K, Liu M, Huang Y*. 2019. Population genomics insights into adaptive evolution and ecological differentiation in streptomycetes. Appl Environ Microbiol, 85: e02555-18. https://doi.org/10.1128/AEM.02555-18
8. Song F, Liu N, Liu M, Chen Y, Huang Y*. 2018. Identification and characterization of mycemycin biosynthetic gene clusters in Streptomyces olivaceus FXJ8.012 and Streptomyces sp. FXJ1.235. Mar Drugs, 16: 98.
9. Zhang L, Dong H*, Liu Y, Bian L, Wang X, Zhou Z, Huang Y*. 2018. Bioleaching of rare earth elements from bastnaesite-bearing rock by actinobacteria. Chem Geol, 483: 544-557. https://doi.org/10.1016/j.chemgeo.2018.03.023
10. Zhang L, Xi L, Ruan J, Huang Y*. 2017. Kocuria oceani sp. nov, isolated from a deep-sea hydrothermal plume. Int J Syst Evol Microbiol, 67: 164-169.
11. Chen P, Zhang L, Guo X, Dai X, Liu L, Xi L, Wang J, Song L, Wang Y, Zhu Y, Huang L, Huang Y*. 2016. Diversity, biogeography, and biodegradation potential of actinobacteria in the deep-sea sediments along the Southwest Indian Ridge. Front Microbiol, 7: 1340. https://doi.org/10.3389/fmicb.2016.01340
12. Cheng K, Rong X*, Huang Y*. 2016. Widespread interspecies homologous recombination reveals reticulate evolution within the genus Streptomyces. Mol Phylogenet Evol, 102: 246-254. http://dx.doi.org/10.1016/j.ympev.2016.06.004
13. Liu N#, Song L#, Liu M#, Shang F, Anderson Z, Fox DJ, Challis GL*, Huang Y*. 2016. Unique post-translational oxime formation in the biosynthesis of the azolemycin complex of novel ribosomal peptides from Streptomyces sp. FXJ1.264. Chem Sci, 7(1): 482-488. https://doi.org/10.1039/c5sc03021h
14. Liu M#, Liu N#, Shang F, Huang Y*. 2016. Activation and identification of NC-1, a cryptic cyclodepsipeptide from red soil-derivedStreptomycessp. FXJ1.172. Eur J Org Chem, 23: 3943-3948. https://doi.org/10.1002/ejoc.201600297.
15. Liu N#, Song F#, Shang F, Huang Y*. 2015. Mycemycins A-E, new dibenzoxazepinones isolated from two different streptomycetes. Mar Drugs, 13(10): 6247-6258. https://doi.org/10.3390/md13106247
16. Huang B, Liu N, Rong X, Ruan J, Huang Y*. 2015. Effects of simulated microgravity and spaceflight on morphological differentiation and secondary metabolism of Streptomyces coelicolor A3(2). Appl Microbiol Biotechnol, 99(10): 4409-4422. https://doi.org/10.1007/s00253-015-6386-7
17. Guo X#, Liu N#, Li X, Ding Y, Shang F, Gao Y, Ruan J, Huang Y*. 2015. Red soils harbour diverse culturable actinomycetes that are promising sources of novel secondary metabolites. Appl Environ Microbiol, 81: 3086-3103. https://doi.org/10.1128/AEM.03859-14
18. Cheng K#, Rong X#, Pinto-Tomás A, Fernández-Villalobos M, Murillo-Cruz C, Huang Y*. 2015. Population genetic analysis of Streptomyces albidoflavus reveals habitat barriers to homologous recombination in the diversification of streptomycetes. Appl Environ Microbiol, 81: 966-975. https://doi.org/10.1128/AEM.02925-14
19. Rong X, Doroghazi JR, Cheng K, Zhang L. Buckley DH, Huang Y*. 2013. Classification of Streptomyces phylogroup pratensis (Doroghazi and Buckley, 2010) based on genetic and phenotypic evidence, and proposal of Streptomyces pratensis sp. nov. Syst Appl Microbiol, 36: 401-407.
20. Liu N, Shang F, Xi L, Huang Y*. 2013. Tetroazolemycins A and B, two new oxazole-thiazole siderophores from deep-sea Streptomyces olivaceus FXJ8.012. Mar Drugs, 11: 1524-1533.
21. Rong X, Huang Y*. 2012. Taxonomic evaluation of the Streptomyces hygroscopicus clade using multilocus sequence analysis and DNA-DNA hybridization, validating the MLSA scheme for systematics of the whole genus. Syst Appl Microbiol, 35:7-18.
22. Wang H#, Liu N#, Xi L, Rong X. Ruan J, Huang Y*. 2011. Genetic screening strategy for rapid access to polyether ionophore producers and products in actinomycetes. Appl Environ Microbiol, 77: 3433-3442.
23. Rong X, Liu N, Ruan J, Huang Y*. 2010. Multilocus sequence analysis of Streptomyces griseus isolates delineating intraspecific diversity in terms of both taxonomy and biosynthetic potential. Antonie van Leeuwenhoek, 98: 237-248.
24. Rong X, Huang Y*. 2010. Taxonomic evaluation of the Streptomyces griseus clade using multilocus sequence analysis and DNA-DNA hybridization, with proposal to combine 29 species and three subspecies as 11 genomic species. Int J Syst Evol Microbiol, 60: 696-703.
25. Gao P, Huang Y*. 2009. Detection, distribution, and organohalogen compound discovery implications of the reduced flavin adenine dinucleotide-dependent halogenase gene in major filamentous actinomycete taxonomic groups. Appl Environ Microbiol, 75: 4813-4820.
26. Rong X, Guo Y, Huang Y.* 2009. Proposal to reclassify the Streptomyces albidoflavus clade on the basis of multilocus sequence analysis and DNA-DNA hybridization, and taxonomic elucidation of Streptomyces griseus subsp. solvifaciens. Syst Appl Microbiol, 32: 314-322.
27. Qiu D, Ruan J, Huang Y.* 2008. Selective isolation and rapid identification of members of the genus Micromonospora. Appl Environ Microbiol, 74: 5593-5597.
28. Guo Y, Zheng W, Rong X, Huang Y.* 2008. A multilocus phylogeny of Streptomyces griseus 16S rRNA gene clade: use of multilocus sequence analysis for streptomycete systematics. Int J Syst Evol Microbiol, 58: 149-159.
黄英, 张利敏, 郗丽君, 宋磊. 一株深海放线菌及其应用. 授权号:XL201410534724.4, 授权日期: 2017.06.09
黄英, 刘宁, 郗丽君, 张利敏. 一株海洋链霉菌及其应用. 授权号:ZL201110140088.3, 授权日期: 2012.12.19
黄英, 刘宁, 郗丽君, 张利敏. 海洋链霉菌产生的化合物及其应用. 授权号:ZL201110174314X, 授权日期: 2013.03.20
黄英课题组